Active Shape Model (ASM) and Active Appearance Model (AAM)
This is an example of the basic Active Shape Model (ASM) and also the Active Appearance Model (AAM) as introduced by Cootes and Taylor, 2D and 3D with multi-resolution approach, color image support and improved edge finding method. Very useful for automatic segmentation and recognition of biomedical objects.
.
Basic idea ASM:
The ASM model is trained from manually drawn contours (surfaces in 3D) in training images. The ASM model finds the main variations in the training data using Principal Component Analysis (PCA), which enables the model to automatically recognize if a contour is a possible/good object contour. Also the ASM modes contains matrices describing the texture of the lines perpendicular to the control point, these are used to correct the positions in the search step.
After creating the ASM model, an initial contour is deformed by finding the best texture match for the control points. This is an iterative process, in which the movement of the control points is limited by what the ASM model recognizes from the training data as a "normal" object contour.
.
Basic idea AAM:
PCA is used to find the mean shape and main variations of the training data to the mean shape. After finding the Shape Model, all training data objects are deformed to the main shape, and the pixels converted to vectors. Then PCA is used to find the mean appearance (intensities), and variances of the appearance in the training set.
Both the Shape and Appearance Model are combined with PCA to one AAM-model.
By displacing the parameters in the training set with a know amount, an model can be created which gives the optimal parameter update for a certain difference in model-intensities and normal image intensities. This model is used in the search stage.
.
Literature:
- Ginneken B. et al. "Active Shape Model Segmentation with Optimal Features", IEEE Transactions on Medical Imaging 2002.
- T.F. Cootes, G.J Edwards, and C,J. Taylor "Active Appearance Models", Proc. European Conference on Computer Vision 1998
- T.F. Cootes, G.J Edwards, and C,J. Taylor "Active Appearance Models", IEEE Transactions on Pattern Analysis and Machine Intelligence 2001
Examples(2D):
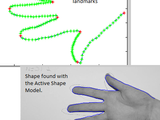
Run the example "ASM_2D_example" and "AAM_AAM_2D_example", the examples are made from manually drawn contours in 10 hand photos (with DrawContourGui). After training the ASM / AAM model will automatically find the contour in the test hand image.
Examples(3D):
First train the classifier to recognize the Mandible "ASM_3D_train_example" or "AAM_3D_train_example", and then segment with "ASM_3D_apply_example", or "AAM_3D_apply_example".
Comments:
Please leave a comment, if you like it, find a bug, or know (made) good improvements to the code.
Cite As
Dirk-Jan Kroon (2024). Active Shape Model (ASM) and Active Appearance Model (AAM) (https://www.mathworks.com/matlabcentral/fileexchange/26706-active-shape-model-asm-and-active-appearance-model-aam), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Image Processing and Computer Vision > Computer Vision Toolbox > Recognition, Object Detection, and Semantic Segmentation >
- AI, Data Science, and Statistics > Statistics and Machine Learning Toolbox > Dimensionality Reduction and Feature Extraction >
- Image Processing and Computer Vision > Image Processing Toolbox > Image Segmentation and Analysis > Image Segmentation >
Tags
Acknowledgements
Inspired: Face detection with Active Shape Models (ASMs), Shape Model Builder
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
AAM Functions/
ASM Functions/
Functions/
InterpFast_version1/
PatchNormals_version1/
PieceWiseLinearWarp_version2/
PieceWiseLinearWarp_version2/functions/
polygon2voxel_version1j/
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.5.0.0 | Many bugfixes and ASM and AAM optimizations |
||
| 1.4.0.0 | Added 3D examples.
|
||
| 1.3.0.0 | Added Color support, new edge finding method in ASM, and contour starting point GUI. |
||
| 1.2.0.0 | Added the Active Appearance Model |
||
| 1.1.0.0 | PCA bug fix. |
||
| 1.0.0.0 |